Protein analysis: Less is more
CONAN to the rescue! The new software-package for molecular dynamic simulations compresses 3D data to contact maps and helps to analyze protein structures. The tool CONAN (CONtact ANalysis), developed at HITS, has now been presented in the latest issue of „Biophysical Journal“.
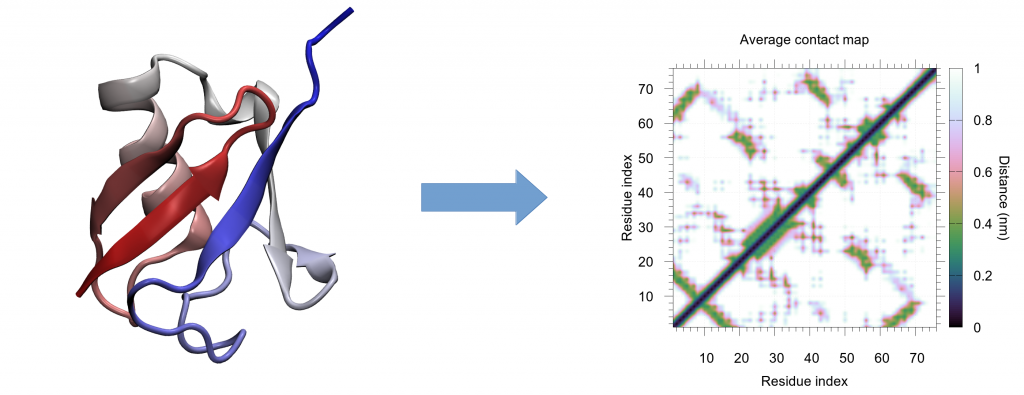
Proteins constantly move and change their conformation. Molecular dynamics typically answers the question of what the possible conformations of proteins are. Proteins, however, have a highly complicated and crowded structure, and understanding the changes in their behavior is a challenging task due to the high number of coordinates to monitor. Digesting the large amount of molecular data often involves creative 3D visualization, but even with considerable effort, important details can be missed. This led to a dual problem; not only was data visualization a challenge, but scientists also ran the risk of overlooking aspects of their own results. A novel tool called CONAN (CONtact ANalysis), developed from the Molecular Biomechanics group at HITS, can alleviate these issues through compressing this 3D data into simpler 2D images capturing the key interactions, named contact maps.
Contact maps measure inter-residue distances, thereby compressing 3D structures into 2D images. This often facilitates data interpretation and makes important changes easier to spot. These contact maps have usually only been used to study single protein structures as a single snapshot, but in fact they can easily be obtained for many structures, resulting in a contact map movie. This analysis somehow extends the saying “a figure is worth more than 1000 words” into the dynamic regime, since it creates a multitude of possible contact-map snapshots out of one simulation, identifying conformational subpopulations and transitions.
Until now, contact maps-based analysis methods have been widely used only as understanding single structures, such as those in the protein data base (PDB). Even when the methods were generalized for dynamic simulations, the implementations were often various “ad hoc” analysis scripts, since there wasn’t a standardized tool. This meant that the measured quantities and definitions were inconsistent and results weren’t directly comparable. The new tool “CONAN” however is a standardized, easy-to-use package that allows several different types of analyses, for example including principal component analysis and cluster analysis. The tool developed by the HITS researchers Csaba Daday and Frauke Gräter of the Molecular Biomechanics group as well as former group member Davide Mercadante therefore fills a gap and offers a comprehensive, user-friendly program requiring no programming experience that can help scientists performing molecular dynamics calculations understand and present their data. Hopefully, this will lead to a more widespread use of these measures, and a more uniform set of definitions. The tool is open access and free of use. The team at HITS also constantly optimizes the software and is open to feedback from the community.
CONAN is available online here.
Examples and illustrations can be found on our blog and our YouTube channel
Article in Biophysical Journal:
CONAN: A Tool to Decode Dynamical Information from Molecular Interaction Maps. Davide Mercadante, Frauke Gräter, Csaba Daday. Biophysical Journal,
Volume 114, Issue 6, p1267–1273, 27 March 2018. DOI: https://doi.org/10.1016/j.bpj.2018.01.033
Scientific Contact:
Prof. Dr. Frauke Gräter
Group Leader „Molecular Biomechanics“
HITS – Heidelberg Institute for Theoretical Studies
E-mail: frauke.graeter@h-its.org
Dr. Csaba Daday
Group Member „Molecular Biomechanics“
HITS – Heidelberg Institute for Theoretical Studies
E-mail: Csaba.Daday@h-its.org
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.