Making sense of what you see in biomedical images
Sometimes an image is just an image. Sometimes it gives those who can read it correctly a deeper insight into what they can see. In many scientific disciplines, the key to extracting meaningful information from large three-dimensional images, obtained from X-ray tomography or optical microscopy, is segmentation, a tedious and time-consuming – and therefore error-prone – task if done manually. An interdisciplinary team of researchers from the Heidelberg Institute for Theoretical Studies (HITS), Heidelberg University, the Karlsruhe Institute of Technology (KIT) and the Technical University of Darmstadt now present Biomedisa, an easy-to-use open-source online platform for biomedical image segmentation. The work, which was published in the journal Nature Communications, addresses the needs of scientists without substantial computational expertise.
Recent years have seen dramatic improvements in imaging technologies that result in higher resolutions and faster acquisition times. Images of single cells, tissue and organs provide medical experts around the world with a myriad of information about their patients’ state of health at a given time. But how do they gain understanding of what they see in these biomedical images?
The status quo: time-consuming and prone to errors
To make these large volumetric images reveal their true information potential, manual segmentation – whereby a digital image is divided into various segments to enable or facilitate analysis – is often required. Labels, such as for example “background” or “object”, are assigned to various structures of interest with different intervals inside the 3D volume. This is followed by an interpolation of the labels between the pre-segmented slices, where values at unknown points are estimated by using known data. In this process, the underlying image data is usually not taken into account, and the interpolation is therefore based exclusively on the segmented slices. Consequently, only a fraction of the real experimental information is utilized to derive the segmentation.
“Manual segmentation of large biomedical datasets of unknown composition is often very time-consuming and prone to errors. For analyzing three-dimensional image data, manual segmentation is still a very common approach. In fact, institutes employ armies of trained students just for this very task,” says Philipp Lösel from the research group “Data Mining and Uncertainty Quantification” (DMQ) at HITS, who developed Biomedisa.
Biomedisa: faster, user-friendly and and more accurate
And this is where the Biomedical Image Segmentation App Biomedisa (https://biomedisa.org) comes in, a free and easy-to-use open-source online platform especially developed for semi-automatic segmentation. The segmentation is based on a smart interpolation of sparsely pre-segmented slices taking into account the complete underlying image data. This makes Biomedisa particularly valuable when little a priori knowledge is available. “Biomedisa can accelerate the segmentation process enormously, while at the same time providing more accurate results than the manual segmentation,” says Thomas van de Kamp (KIT), a biologist with painful experience in manual image segmentation who provided micro-CT data and evaluated Biomedisa during its development.
The platform is accessible through a web browser and requires no complex and tedious configuration of software and model parameters. The one-button solution can be used for different 3D imaging modalities and various biomedical applications.
“Our explicit aim,” summarizes Vincent Heuveline, director of Heidelberg University’s Computing Centre (URZ) and DMQ group leader at HITS, “was to create a widely-applicable and user-friendly tool to accelerate the segmentation of samples of unknown morphology while also improving the results.”
“Biomedisa is an example for a software that benefits directly from the latest developments of GPU (Graphics Processing Unit) technology. The hardware-aware design utilizes graphics accelerators to handle the ever-increasing image data,” adds Philipp Lösel.
On the way to fully automatic segmentation
Besides, Biomedisa offers a range of other functions, such as the removal of outliers or the filling of holes, surfaces can be smoothed and the uncertainty with which the result was obtained can be quantified. Furthermore, the data can be visualized with 3D rendering software and shared with other users.
Last but not least, Biomedisa enables machine learning techniques through training a deep neural network. This technique allows a fully automatic segmentation when a large number of similar structures, such as the human heart, is segmented. As a result, it allows numerical simulations based on a patient-specific heart model and thus assists clinicians with their surgical planning and decision-making.
All these features combined make Biomedisa an ideal platform for all those for whom a picture is worth more than a thousand words.
Philipp D. Lösel, Thomas van de Kamp, Alejandra Jayme, Alexey Ershov, Tomáš Faragó, Olaf Pichler, Nicholas Tan Jerome, Narendar Aadepu, Sabine Bremer, Suren A. Chilingaryan, Michael Heethoff, Andreas Kopmann, Janes Odar, Sebastian Schmelzle, Marcus Zuber, Joachim Wittbrodt, Tilo Baumbach & Vincent Heuveline: Introducing Biomedisa as an open-source online platform for biomedical image segmentation. Nature Communications, 4 November 2020. DOI 10.1038/s41467-020-19303-w
https://www.nature.com/articles/s41467-020-19303-w
Press Contact:
Dr. Peter Saueressig
Head of Communications
Heidelberg Institute for Theoretical Studies (HITS)
Phone: +49-6221-533-245
peter.saueressig@h-its.org
Scientific Contact:
Philipp Lösel
Data Mining and Uncertainty Quantification group (DMQ)
Heidelberg Institute for Theoretical Studies (HITS)
Phone: +49-6221-533-520
philipp.loesel@h-its.org
Biomedisa Tutorials on YouTube
https://www.youtube.com/channel/UCTNOthYVKyIWVvYYZSU_mfQ
Image:
Biomedisa Fig. 1
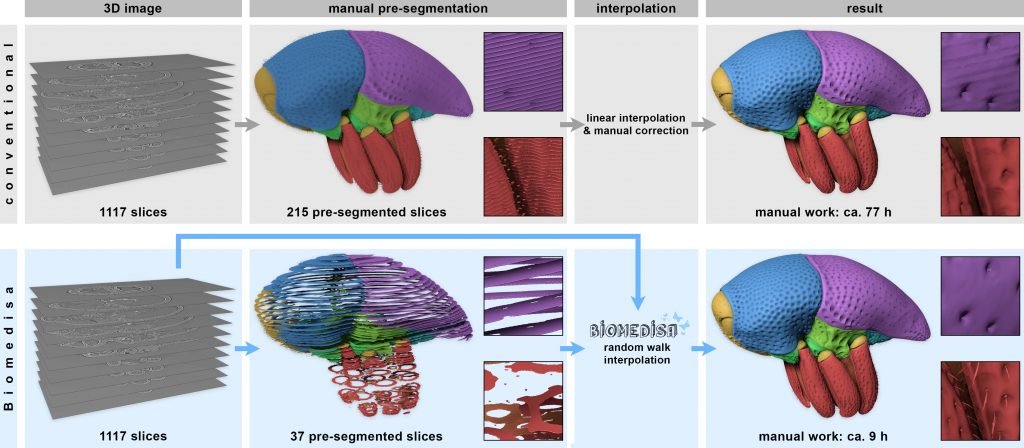
Comparison between a conventional segmentation approach (top row) and Biomedisa (bottom row): The conventional procedure requires 77 hours, compared to 9 hours with Biomedisa. Both procedures require manual pre-segmentation of the 3D image stack. While the widely used morphological interpolation solely considers labels on pre-segmented slices, Biomedisa takes both the underlying 3D image data and the pre-segmented slices into account, resulting in a significantly lower amount of required manual input. Moreover, interpolation artifacts are avoided and fine details like hairs, which are usually omitted during manual segmentation, are included.
(Image: Lösel)
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.