τRAMD – compute protein-ligand dissociation rates and explore dissociation mechanisms
Researchers from the Molecular and Cellular Modeling (MCM) group at HITS have developed and validated τRAMD (τ-Random Acceleration Molecular Dynamics),an efficient computational workflow that enables the prediction of drug-protein relative residence times and the analysis of dissociation mechanisms in an automated manner. As such, τRAMD is a flexible tool that can be implemented in rational drug design workflows for the design of new molecules or ligand optimization.
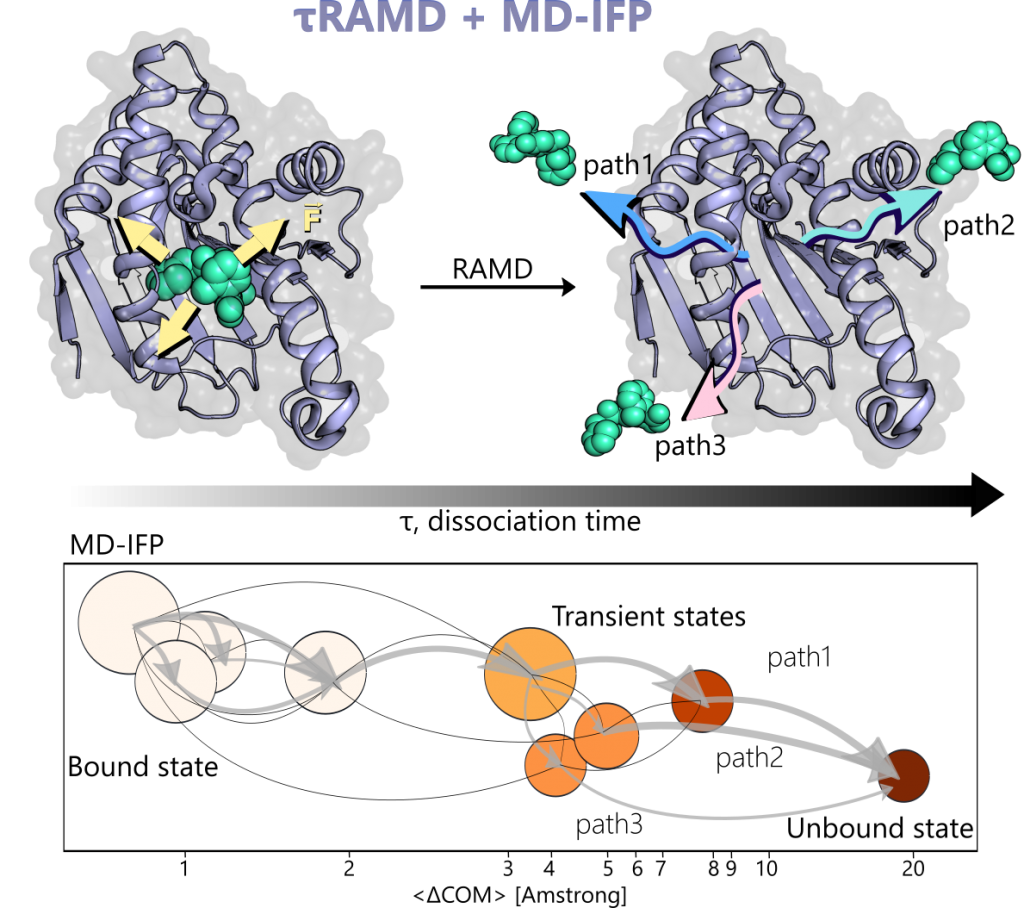
The τRAMD protocol applies RAMD to accelerate the egress of a small molecule from a target receptor through the application of an adaptive randomly oriented force on the ligand. The workflow facilitates the identification of dissociation mechanisms and characterization of transition states through the MD-IFP (Molecular Dynamics – Interaction Fingerprint) post-analysis tool.
In addition to implementations in the NAMD and AMBER software packages, RAMD has recently been implemented in the freely available GROMACS molecular simulation engine for simulations on CPU or GPU nodes.
References:
Kokh DB, Doser B, Richter S, Ormersbach F, Cheng X, Wade RC (2020). A workflow for exploring ligand dissociation from a macromolecule: Efficient random acceleration molecular dynamics simulation and interaction fingerprint analysis of ligand trajectories, J. Chem. Phys. 153(12):125102
Kokh DB, Amaral M, Bomke J, Grädler U, Musil D, Buchstaller H, Dreyer MK, Frech M, Lowinski M, VKokh DB, Amaral M, Bomke J, Grädler U, Musil D, Buchstaller H, Dreyer MK, Frech M, Lowinski M, Vallee F, Bianciotto M, Rak A, Wade RC (2018). Estimation of Drug-Target Residence Times by τ-Random Acceleration Molecular Dynamics Simulations, J. Chem. Theory Comput. 14(7):3859-3869
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.