New version of TRAPP released
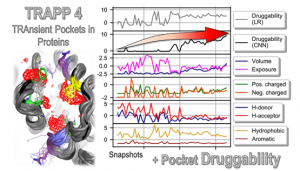
Version 4 uses machine learning and molecular simulation to predict druggability
Researchers from the Molecular and Cellular Modeling (MCM) group at HITS released a new version of the TRAnsient Pockets in Proteins (TRAPP) webserver. Version 4 enables the exploration of changes in the shape of a given binding pocket due to protein motion and of how these changes affect the physicochemical properties of the pocket and ability to bind to molecules like drugs. TRAPP facilitates the discovery of cryptic binding pockets that may not be apparent in the experimentally determined structure of a protein but that can be revealed when the protein motions are simulated.
The main new feature in TRAPP version 4 is the option to score the druggability, i.e. the ability to bind to a drug-like compound, of each conformation (or shape) of the protein binding site studied. The druggability score can help the user to easily identify the most druggable conformation of a given binding pocket from a set of conformations obtained from experiments or a molecular dynamics simulation. This predicted druggable conformation can then be used for drug design, e.g. by docking molecules into this pocket conformation.
Machine learning was used to develop two statistical models for predicting druggability in TRAPP. The models, which were trained on publicly available and self-augmented datasets, show equivalent or superior performance to existing methods on test sets of protein crystal structures and have sufficient sensitivity to identify potentially druggable protein conformations in trajectories from molecular dynamics simulations. TRAPP includes visualization tools that enable the user to identify the factors affecting the predicted druggability of protein binding pockets.
The TRAPP webserver is freely available for use by the scientific community.
References:
TRAPP webserver: https://trapp.h-its.org/
Jui-Hung Yuan, Sungho Bosco Han, Stefan Richter, Rebecca C Wade, Daria B. Kokh
Druggability Assessment in TRAPP using Machine Learning Approaches
J. Chem. Inf. Model. (2020) 60(3):1685-1699, DOI: 10.1021/acs.jcim.9b01185
Preprint on BioRxiv: doi: 10.1101/2019.12.19.882340
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.