It’s in the DNA: sequence recognition by the linker histone
Using single-pair FRET (Fluorescence/Förster Resonance Energy Transfer) spectroscopy and computational modeling, Madhura De, researcher in the Molecular and Cellular Modeling (MCM) group at HITS, has shown how certain DNA sequences orient the linker histone (also known as H1) on the nucleosome. In particular, this study, now published in Biophysical Journal, sheds light on how the linker histone recognizes stretches of contiguous adenines (so-called adenine or A-tracts) on the DNA.
The human DNA present in a single cell is about 2 meters long when fully unraveled. However, it fits in a cell nucleus which is only a few micrometers across. The key to fitting all the DNA into such a small volume is an elaborate packaging system. In a first step, the DNA is wrapped around core histone proteins forming so-called nucleosomes. A string of nucleosomes then forms chromatin. One of the proteins that enables this efficient packaging mechanism is the linker histone. Linker histones bind on the outer side of the nucleosome right at the juncture where the DNA arms (so-called linker-DNA) come out of the nucleosome (see figure). Being highly positively charged, the linker histone effectively neutralizes the negative charges on the DNA and thereby compacts the nucleosome.
Linker histones are ubiquitous. But previous research has shown that the linker histone prefers AT-rich DNA sequences and A-tracts. However, it was unclear why they favor this base pair over others and how they recognize particular sequences in the first place. The answer to this question might help to understand how the linker histone is able to compact certain AT- and A-tract (runs of 4-6 adenine base pairs) rich regions of the DNA, such as found in the highly compacted heterochromatin, and scaffold associated regions.
A dual approach
The published paper focuses on exactly this question by comparing different DNA sequences to understand what influences the position of the linker histone on the nucleosome. For her research, HITS scientist Madhura De used a dual approach of experiments and modeling and analyzed the distance of the linker histone to the two emerging linker-DNA arms for several DNA sequences. By using FRET spectroscopy (FRET = Fluorescence/ Förster Resonance Energy Transfer), a biophysical technique commonly used to measure distances on a biomolecular level using fluorescent dyes, she found out that the linker histone is positioned in a particular orientation. The data confirmed that the linker histone is closer to the linker-DNA arm that contained an A-tract. Her research results suggest that the linker histone does not recognize this DNA sequence directly, but rather through the geometric parameters of the DNA that depend on sequence. The study revealed a new recognition mechanism for how linker histones can recognize adenine-tracts, using conserved arginine residues binding in the DNA minor groove, similarly to the mode of recognition that has previously been identified in other A-tract recognizing proteins. The results of this study thus help to understand the linker histone’s preferences for adenine tracts which are abundantly found in certain parts of the genome, such as the scaffold associated region and heterochromatin, and have been found in oncogenes.
HITS researcher Madhura De conducted the experiments for this study at the German Cancer Research Center (DKFZ) and did the computational modeling and analysis at HITS. Additionally, this paper uses an efficient FRET-data processing software developed by coauthor Dr. Sebastian Isbaner (currently at IMP Vienna, Austria): AlexEval, which is available online: https://github.com/sisbaner/AlexEval
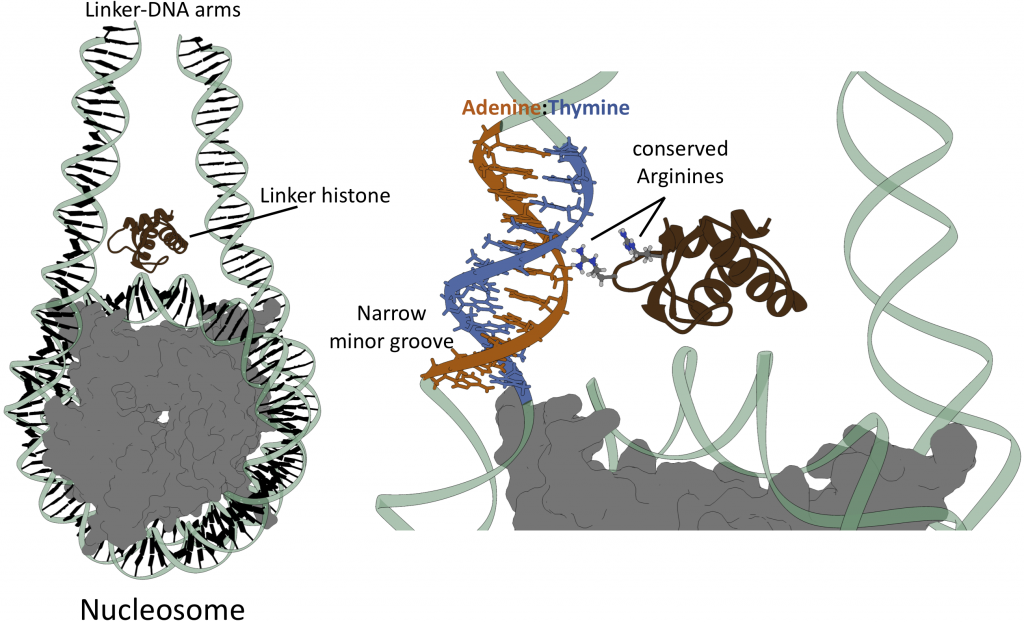
A nucleosome, the smallest repeat unit of chromatin, is shown on the left (DNA in pale green and core histones in grey), with the linker histone (dark brown) associated with it. The close-up view on the right illustrates the recognition mechanism revealed in this study in which two arginine residues on the linker histone approach the minor groove of an A(adenine)-tract (adenine in orange, thymine in blue). Picture: Madhura De, HITS
Publication
DNA sequence-dependent positioning of the linker histone in a nucleosome: A single-pair FRET study
Madhura De, Mehmet Ali Öztürk, Sebastian Isbaner, Katalin Tóth, Rebecca C. Wade
Published: July 19, 2021
DOI: https://doi.org/10.1016/j.bpj.2021.07.012 https://www.cell.com/biophysj/fulltext/S0006-3495(21)00596-8
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.