Predicting the Ever-Changing World of Protein Dynamics
In July 2025, a team of researchers from HITS and the Max Planck Institute for Polymer Research (MPIP) developed a model that learns how to generate proteins with highly flexible structures. A new, complementary deep learning model — Back Bone Flow (BBFlow) — can now predict the dynamics of proteins, providing insights into how they function. Their work was showcased at international conferences in San Diego and Copenhagen in December 2025.
Unlike the existing models GAFL (Geometric Algebra Flow Matching) and FliPS (Flexibility-Conditioned Protein Structure Design with Flow Matching), this flow-based deep learning model can capture a protein’s three-dimensional structure using only the geometry of the backbone — the spatial arrangement of a protein’s main chain of atoms. Instead of relying on complex simulations or evolutionary sequence data, the geometry of the protein backbone alone is sufficient.
“While BBFlow predicts protein dynamics, FliPS designs new proteins with specific dynamic properties. BBFlow can then filter the best candidates generated by FliPS,” says Nicolas Wolf, first author of the paper.
Furthermore, BBFlow is accessible to everyone, intuitive to use without any ML coding knowledge, and runs up to 40 times faster than the state-of-the-art AlphaFlow model with comparable accuracy. Unlike conventional methods that rely on evolutionary sequence information, BBFlow does not require such data. It predicts protein dynamics directly from their structure, making it particularly well-suited for new or synthetic proteins that lack natural counterparts.
Fast, Efficient, and Accurate: A Dance of Endless Conformations
It is the high speed, in particular, that gives BBFlow its primary appeal. The model can be trained “from scratch” in just a few days, that is, without extensive pretraining or evolutionary information. Tests showed it produces realistic ensembles for both naturally occurring proteins and newly designed (de novo) proteins, which typically lack evolutionary data.
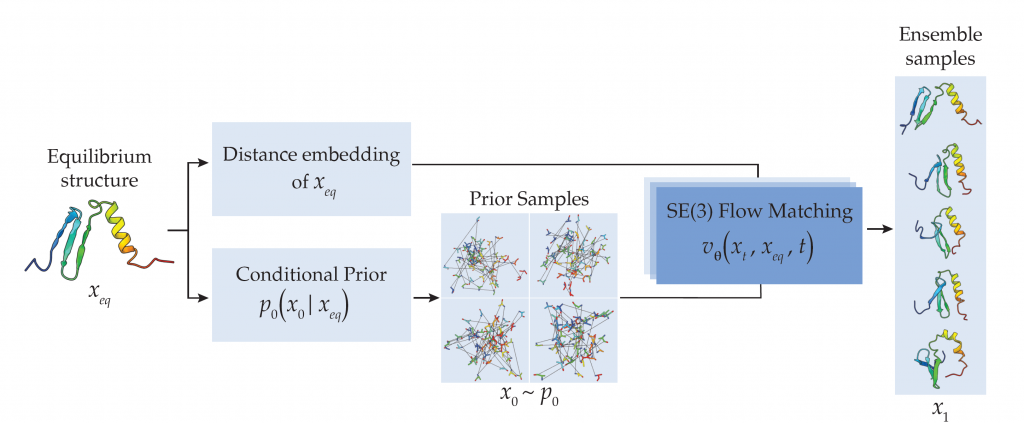
“Rather than relying on evolutionary data, we use a method that directly generates protein structures. The model is guided by the protein’s known starting structure,” says Leif Seute, a PhD student in the Machine Learning and Artificial Intelligence (MLI) group and co-author of the study. This leads to much faster predictions while maintaining comparable accuracy. “The approach works not only for natural monomeric proteins, but also for de novo and multimeric proteins,” Seute adds.
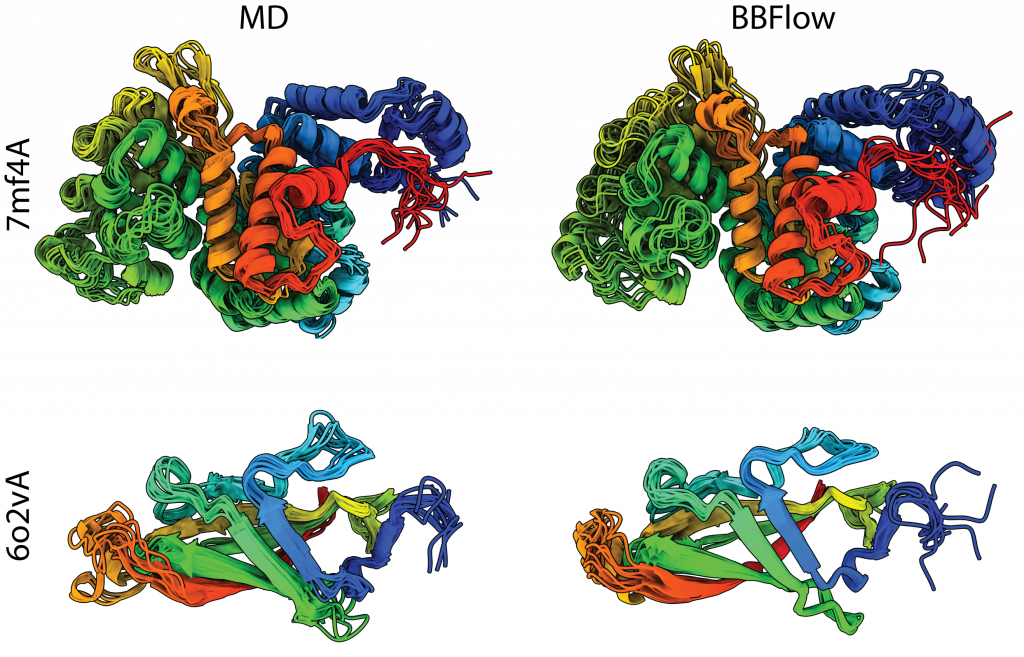
This approach could significantly accelerate protein research and the design of novel proteins which is crucial for developing new drugs, therapies, and materials and where evolutionary comparison data is often missing. This model provides not just a single “functional shape” but a whole ensemble of structures, offering a more realistic view of protein flexibility.
A Step Toward Realistic Protein Models
By shifting the focus from rigid single structures to dynamic ensembles, the teams move closer to capturing how proteins actually behave in the cell. While the current model focuses only on the backbone and does not yet include side chains or full atomic details, it demonstrates that backbone geometry alone carries far more information than previously thought. It opens a new perspective where accuracy, speed, and flexibility in protein design are no longer mutually exclusive.
N. Wolf, L. Seute, V. Viliuga, S. Wagner, J. Stühmer, F. Gräter. Learning conformational ensembles of proteins based on backbone geometry. NeurIPS, 2025.
Scientific Contact:
Jun.-Prof. Dr. Jan Stühmer
Junior Group Leader
Machine Learning and Artificial Intelligence
Heidelberg Institute for Theoretical Studies (HITS)
https://www.h-its.org/people/dr-jan-stuhmer/
Prof. Dr. Frauke Gräter
Director, Head of the Department “Biomolecular Mechanics”
Max Planck Institute for Polymer Research (MPIP)
https://www.mpip-mainz.mpg.de/1001480/01_Direktor
Media contact:
Angela Michel
Head of Communications
Heidelberg Institute for Theoretical Studies (HITS)
+49 (0)6221 533 277
angela.michel@h-its.org
Teresa Petry
Communication
Max Planck Institute for Polymer Research (MPIP)
+49 (0)6131 379-119
pr@mpip-mainz.mpg.de
About HITS
HITS, the Heidelberg Institute for Theoretical Studies, was established in 2010 by physicist and SAP co-founder Klaus Tschira (1940-2015) and the Klaus Tschira Foundation as a private, non-profit research institute. HITS conducts basic research in the natural, mathematical, and computer sciences. Major research directions include complex simulations across scales, making sense of data, and enabling science via computational research. Application areas range from molecular biology to astrophysics. An essential characteristic of the Institute is interdisciplinarity, implemented in numerous cross-group and cross-disciplinary projects. The base funding of HITS is provided by the Klaus Tschira Foundation.