Project 7: Using invertible neural networks to predict molecular interactions
Protein-biomolecule-interactions are ubiquitous in life, from DNA-replication to regulating the heartbeat. The stability of biomolecular complexes depends heavily on the involved proteins’ amino acid sequence.
Changes in this sequence and their effects on complex formation are of interest in many diseases, but also in protein design.
However, predicting how two molecules will interact remains a challenge due to the high degrees of freedom both within and between the interaction partners.
In the recent past, deep neural networks have proved useful for structure prediction of biomolecules.
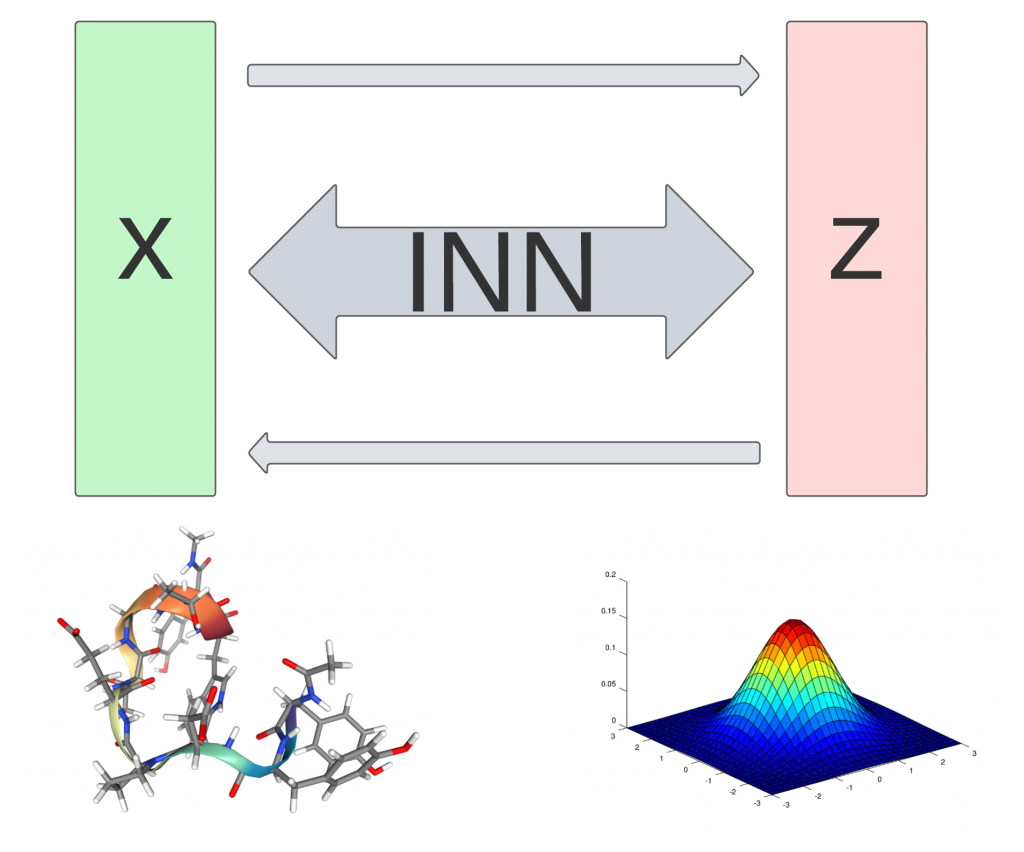
We investigate the suitability of invertible neural networks (INNs) for predicting the structures of molecular complexes, as well as the effects of mutations in sequence. INNs are interesting candidates because they model the full probability distribution of molecular interactions. Also, they do not suffer from the curse of high dimensionality and can therefore be scaled to very large problems like protein-protein-interactions. Major tasks at hand are finding the right representation of the molecular structure and building suitable network architectures. Once we can reliably predict molecular interactions, we will integrate sequence information to learn how sequence alterations influence the structure of biomolecular complexes.
Team

Prof. Dr. Ullrich Köthe
Principal Investigator (IWR, Heidelberg University)
Phone: +49 6221 5414834
More Information